Abstract
Background: The combination of immunomodulatory (IMiDs) drugs with monoclonal antibodies targeting the transmembrane glycoprotein CD38 induces deep and durable responses in MM patients. IMiDs mediated degradation of the transcription factors IKZF1/3 attenuates myeloma cells proliferation and releases IKZF1 transcriptional repression of IL-2 and type I/II interferon response genes promoting T, NK and NKT cells proliferation and activation. Similarly targeting the non-lineage restricted molecule CD38 with daratumumab also exerts direct and indirect tumoricidal effects. Mechanisms implicated in daratumumab-mediated killing include a direct apoptotic effect and activation of cytotoxic immune effector functions, including antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis, and complement dependent cytotoxicity (CDC). Therefore, the synergistic effects observed with daratumumab and IMiDs are postulated to derive from their co-modulation of the host adaptive and innate immunity. A systematic unsupervised interrogation of the bone marrow immune cells of daratumumab and IMiDs treated MM patients is key for the prediction of their clinical responses and the understanding of underlying mechanisms of resistance.
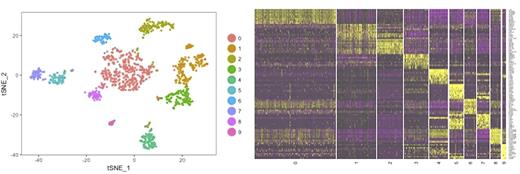
Methods and Results: Bone marrow (BM) aspirates and matching peripheral blood were collected from 20 myeloma patients treated with daratumumab in combination with IMiDs (lenalidomide or pomalidomide) prior to initiation of therapy and at the time of disease response (n=10) or progression (n=10). Bone marrow mononuclear cells were isolated through Ficoll density gradients coupled with magnetic sorting of CD138pos and CD138neg cells. Unbiased mRNA profiling of BM CD138neg cells was performed by single-cell RNA-seq (scRNA-seq) using the GemCode system (10x Genomics), a droplet-based method enabling high cell throughput and efficient single-cell capture. Sequencing was performed on Illumina NEXTseq platform in a paired-end format. Cell Ranger Single Cell Software Suite was used to perform sample de-multiplexing, barcode processing, and single-cell 3′ gene counting. Sequencing data from 30,074 CD138neg single cells (12,908 cells from responding (S) and 17,166 cells from progressing (R) patients) were kept for analysis by the Seurat R package and analyzed by principal component analysis (PCA) and clusteringand then visualized by t -distributed stochastic neighbour embedding (t -SNE) projection. Shown in Fig 1 a representative t -SNE and heatmap plot of 9 immune clusters typically identified in individual samples including terminal effector T cells (KLRG1hi, IL7Rlow), memory T cells (central memory TCM: SELLhi CCR7hi and effector memory TEM: SELLlow CCR7low), CD4 T cells, NK and NKT cells (NKG7, GNLY), B cells (MS4A1), monocytes (CD14, LYZ), macrophages (CD68) and dendritic cells (FCER1A, CST3). Transcriptome profiling of immune cells pre- and post-treatments in S and R patients revealed the following key findings: 1) T cells (CD8Apos and CD3pos) were largely senescent in myeloma patients lacking CD28 expression; 2) amongst checkpoint inhibitory genes LAG3 had the highest expression on activated T cells followed by PDCD1 and to a much lesser extent by HAVCR2 and TIGIT; 3) exposure to daratumumab and IMiDs resulted in the expansion of activated effector CD8 cyctotoxic and effector memory T cells and depletion of CD38pos T and B cells; 4) compared to resistant or progressing patients, responders were characterized by higher CD28 expression in T cells, a significantly larger clusters of central memory T cells (TCM: IL7Rpos, SELLpos, CCR7pos, CD27pos, KLRG1neg) and a M1 activated macrophage signature (IL1Bpos, CCL3pos). Of note no significant difference was noted in CD38 transcript amongst S and R patients. Single cell profiling of CD138pos cells will be updated at the meeting as well as further characterization of the immune repertoire and TCR clonality status of R and S patients.
Conclusion: Anunbiased single cell profiling of bone marrow immune cells in MM patients exposed to Daratumumab and IMiDs demonstrated that LAG3 targeting and restoration of co-stimulatory genes expression (CD28) should be studied as means to reverse immunoparesis. Central memory T cells (TCM) expansion and M1 macrophage signature in responders to Daratumumab and IMiDs combinations represent potential biomarkers of response.
Neri: Celgene: Consultancy, Honoraria, Research Funding; Janssen: Consultancy, Honoraria, Research Funding. Jimenez-Zepeda: Janssen: Honoraria; Amgen: Honoraria; Celgene: Honoraria; Takeda: Honoraria. Boise: Eli Lilly and Company: Research Funding; Abbvie: Consultancy. Thakurta: Celgene Corporation: Employment, Equity Ownership. Bahlis: Takeda: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Celgene: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Janssen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Amgen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.